Downloading and Installing R and RStudio
Resources for this project
- R Markdown document called “The (very) basics of RStudio” for this exercise. You’ll learn to use it below.
What is R?
R started as a free, open-source version of the statistical programming language S and S+. However, because it is open-source, many biologists have learned to use it and contributed to it and it is now the industry-standard statistical software. You can use it to do statistical analysis (not surprisingly), draw complex graphs, control other software, analyze DNA sequences and much more.
You may be very resistant to learning to program in a biology class, but biology is no longer an observational and descriptive science – it increasingly incorporates mathematical models and complex experimental design. Therefore, learning to use the tool, R, that has become essentially standard in the field, will benefit you greatly. It won’t come easy for many of you, but it’ll be worth it in the long run. Additionally, you’ll be able to put on your resume that you know a little bit about using it!
Because this is a biology class, we will only learn some very basic things about R. Primarily, we’ll use R to read in data, manipulate it a bit, and then make a graph and/or do a simple statistical test. It is likely that you have used Microsoft Excel in the past, and you will need to get a bit used to a different way of doing things. R doesn’t have a spreadsheet program to type and store data, so we will use Google Sheets for that. We will then tell R to grab the data from Google, manipulate it and output a graph or statistical result. Unlike Excel, we will be doing a bit of real programming in order to tell the computer what to do.
Choosing between RStudio and Posit Cloud
There are 2 convenient ways to use R. I recommend the first option for this class:
- Use this option if you use a Mac or a PC for which you can install software. Do both of the following steps:
- Download and install R.
- Download and install RStudio Desktop. RStudio provides a nice user interface to R. R itself is much more difficult to use without such an interface, and RStudio is very good! Installation should be easy if you use Windows, MacOS, or (standard) Linux. If you use a Chromebook, you won’t be able to install it and will have to use one of the online options.
- Use this option if you use a Chromebook, iPad or if you cannot install software to your computer.
- Use Posit Cloud. This provides full functionality in a convenient web app. To use this option, you’ll need to sign up for a free account. Although this option won’t require you to install anything, you’ll find that getting your files into and out of Posit Cloud more difficult than if you used the stand-alone apps in option 1. The user interface for Posit Cloud and RStudio are identical. You should be able to use Posit Cloud with an iPad, however getting files in and out is much more difficult, and you really should have a keyboard! Please use any computer other than an iPad.
Getting Started
Please follow along by actually doing the steps listed below. You can’t learn a new skill without practicing. Trust me, this will save you lots of frustration later on.
Open up the RStudio application or log on to your Posit Cloud account. You should see your window divided into 3 or 4 sections (Fig. 1).
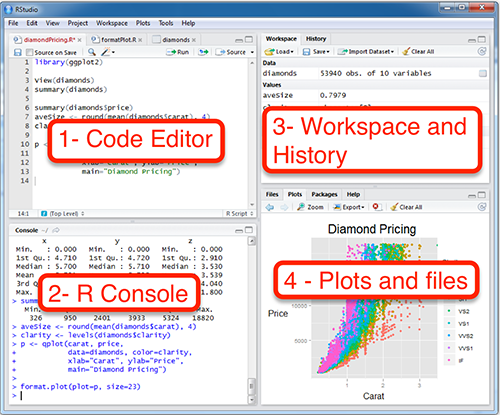
- Code editor. This is where you should write your code and save it.
Rconsole. This is whereRactually does its work. You will usually be able to ignore this section and just observe it for its output. That output will give you information that may help you debug your code.- Workspace and history. You will usually ignore this as well, however it can be useful for making sure that your data is read in properly.
- Plots and files. When you draw a graph, this is where it will appear. This pane can also be used for getting ‘help’ on an
Rcommand. You can also see files in your project here.
R File types
The simplest R files are called “R script” files that list, in order, the commands to execute. We will be using a slightly more complex file called ‘R markdown’ which allows us to mix computer R code with comments and explanations. It also allows us to produce a single finished document with all the figures integrated. These R markdown documents always have a file extension of “.Rmd”
Important note It is possible to run R one command at a time, but it is much better to save your commands in a file. This will allow you to re-run the code easily if you decide to change anything, or if your underlying data changes.
Getting Started – loading an R Markdown file
- Download this file and save it on your computer. I recommend you stay organized by starting a new folder (directory) for Rmd files for this class.
- I highly recommend that you make a new directory (folder) on your computer to store Rmd files for this class. Believe me, this will make your life easier, so don’t skip doing this.
- Whenever you download an analysis file, save it into this newly-created directory and open it from there.
- Use the following steps for every lab in which we use R.
- If you are using a stand-alone version of RStudio
- Go to the file menu, and select “Open File…”
- Find the file where you saved it. It will open in the top-left quadrant.
- Go to the “Session” menu and select “Set Working Directory” and then “To source file location” (Figure 6 above and 7 below.)
- If you are using Posit Cloud
- Go to “Your Workspace” (see red boxes in Figure 2).
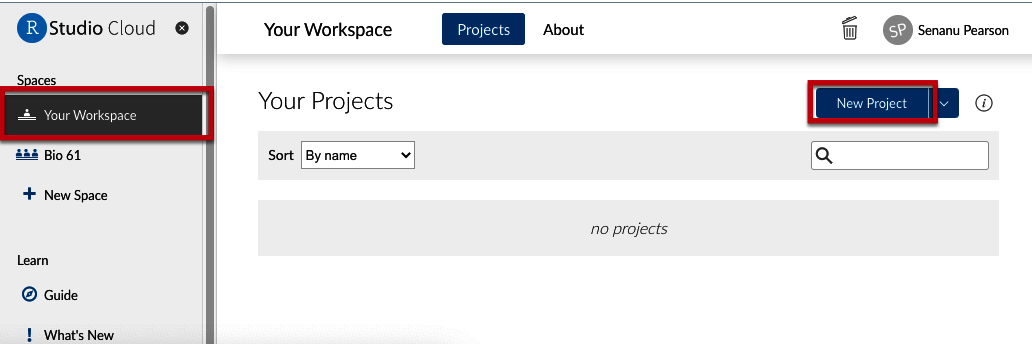
Figure 2. Loading files in Posit Cloud - Click on “New Project”. R will start.
- In the bottom-right quadrant, choose “Upload” and direct Posit Cloud to the file you downloaded and saved on your computer.
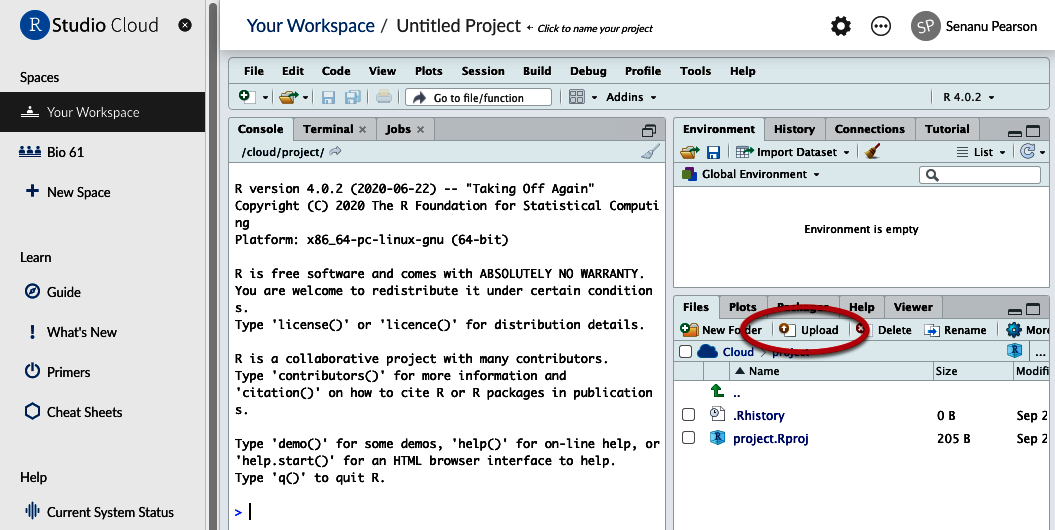
Figure 3. Location of Upload button - You will see that file appear in the list of files. Click on it and it will open in the top-left quadrant.
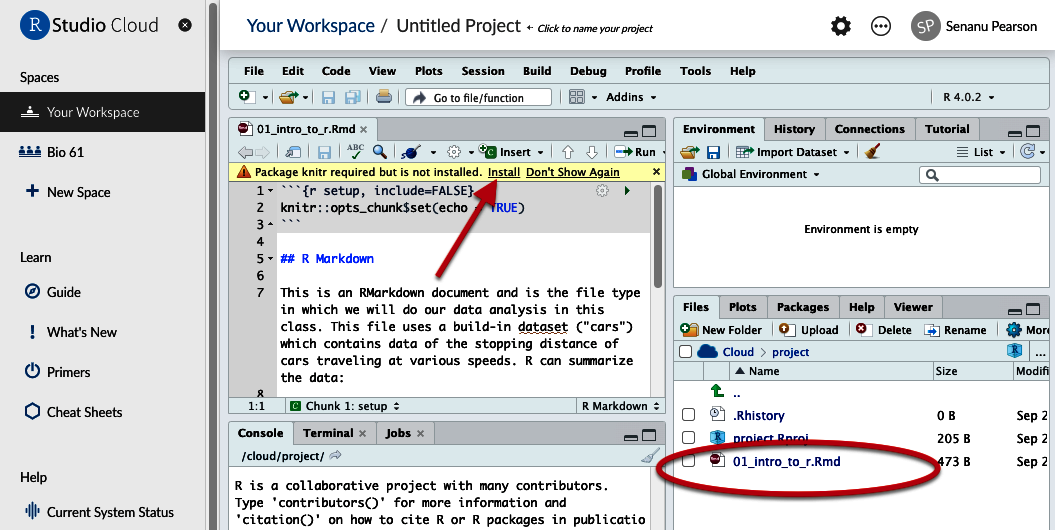
Figure 5. Location of uploaded file and Message to install Knitr - You will probably be given a warning that a package called ‘knitr’ is required but not installed. Click install. It will install on the web server, not on your computer.
- Go to the “Session” menu and select “Set Working Directory” and then “To source file location”.
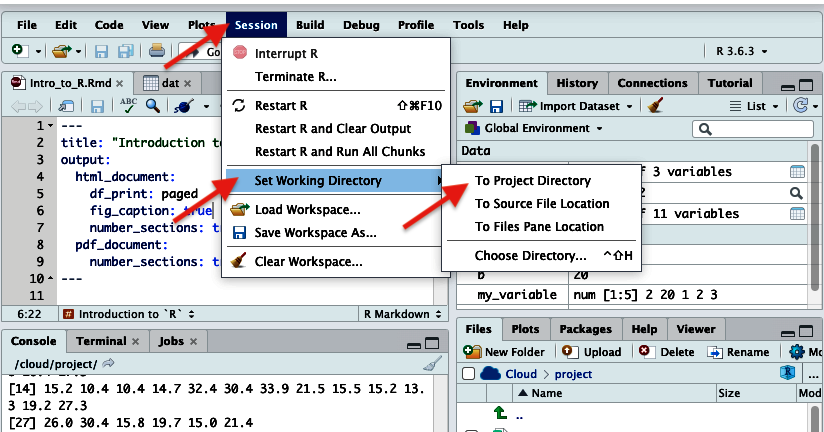
Figure 6. Setting the working directory
- Go to “Your Workspace” (see red boxes in Figure 2).
You will need to do the above steps for every project we do. You should now have the R Markdown file loaded into either RStudio or Posit Cloud. It should look like the attached picture.
Installing packages
R allows users to extend the language by writing ‘packages’ which contain commands for (often) specialized tasks. We will take advantage of a few of those. For now, the main one we need is called ‘knitr’. If you are using Posit Cloud, you probably don’t need to do this, but if you are using RStudio itself, you probably will.
- Depending on the file, you may see a yellow strip across the top indicating that you need to install a package. Simply click “install”
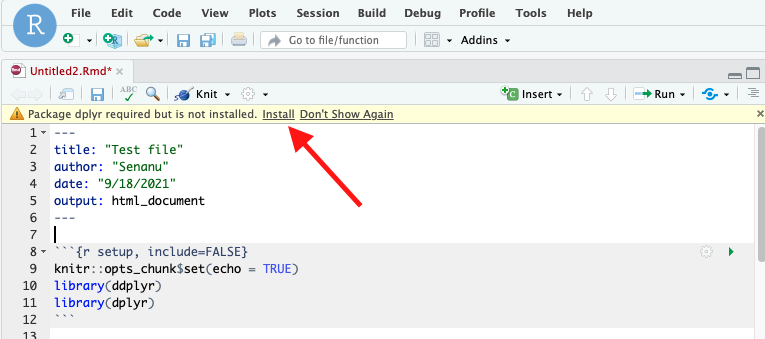
Figure 8. Yellow strip indicating you need to install packages - If you don’t see a yellow strip and you need to install packages, then in the lower-right quadrant, choose the “Packages” tab, then click “Install”.
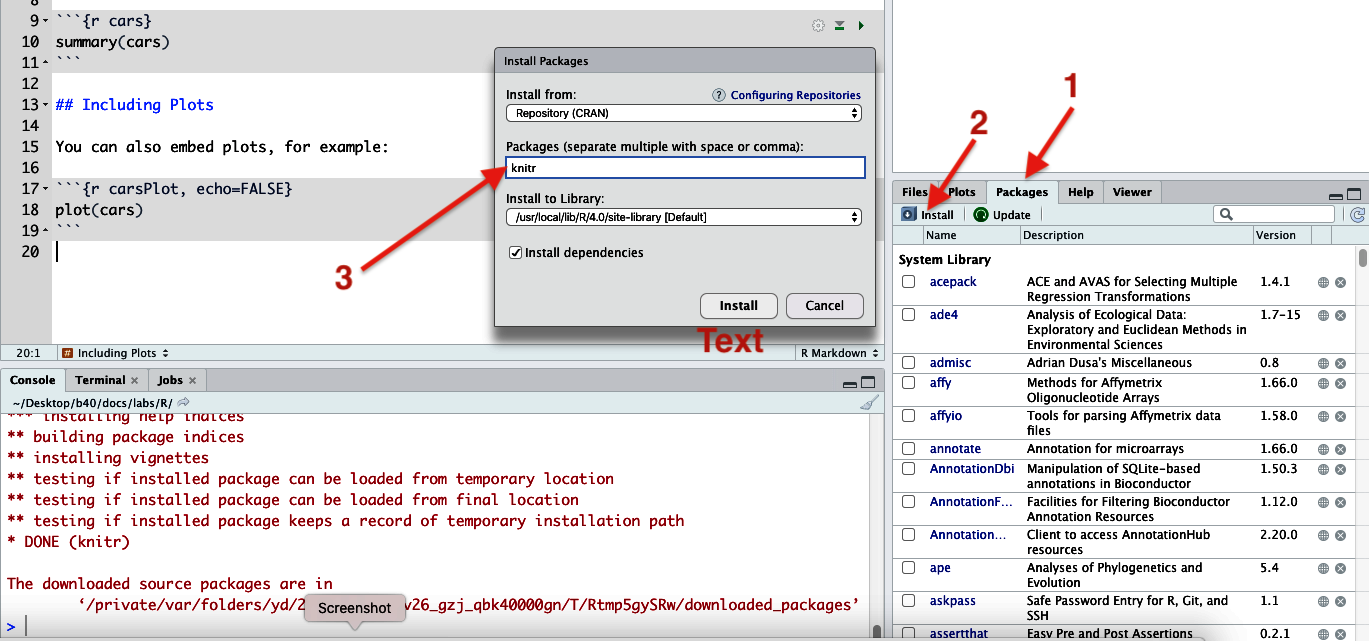
Figure 7. Installing packages (knitr in this case) - In the pop-up dialog box, type “knitr”, then click “Install”
- You will see messages scroll up in the lower-left quadrant. Near the end, it should say “* Done (knitr)”.
Once a package is installed, you shouldn’t have to install it again (unless you do lots of upgrading at a later date)
Using R Markdown files
Now we will look at how to use the R Markdown files. As mentioned above, these files combine code with comments. The comments help us communicate in natural language what we are trying to do with code, in order for another person to be able to understand the code quickly. It is useful to include comments for your own code, because if you look at it a few weeks later, you may struggle to figure out what and why you did what you did.
The gray regions are called ‘code chunks’. These are the actual bits of code that R runs. The white areas are for comments and explanations. There are three main ways in which you can run the code.
- To run all of the code in the entire file, choose “Run” – “Run All” as shown below.
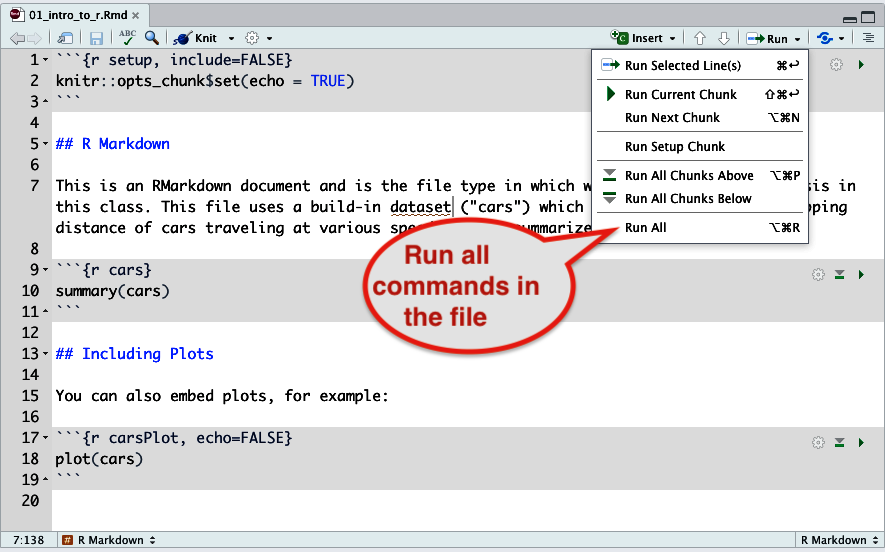
Figure 9. How to run all commands in a file. - You can run individual gray code chunks by clicking the little green arrow.
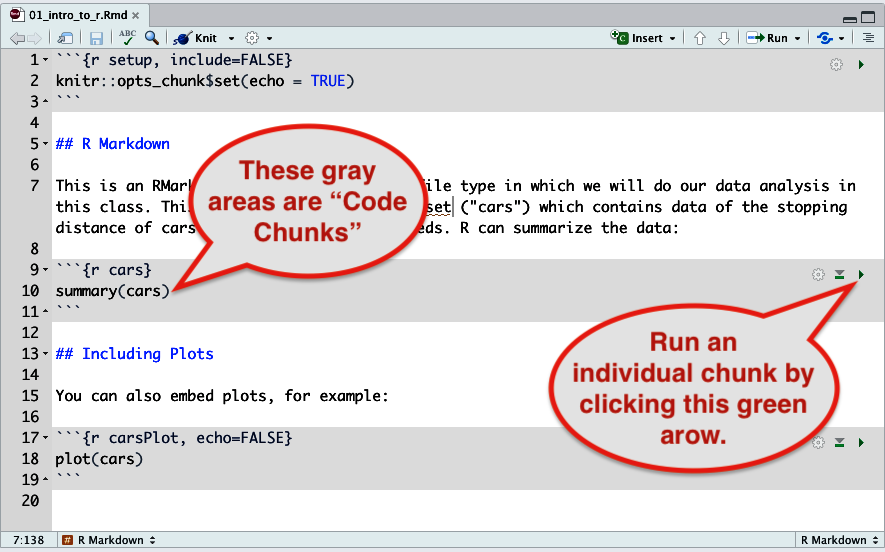
Figure 10. Click the green arrow to run a single code chunk (in gray) - You may run a single line by pressing “control-enter” (on a Windows PC) or “command-enter” (on a Mac).
Please become familiar with each of these 3 methods by using them on the gray parts of the file you have loaded.
You can view the Rmarkdown document in either “Source” or “Visual” mode, controlled by a button near the top left.
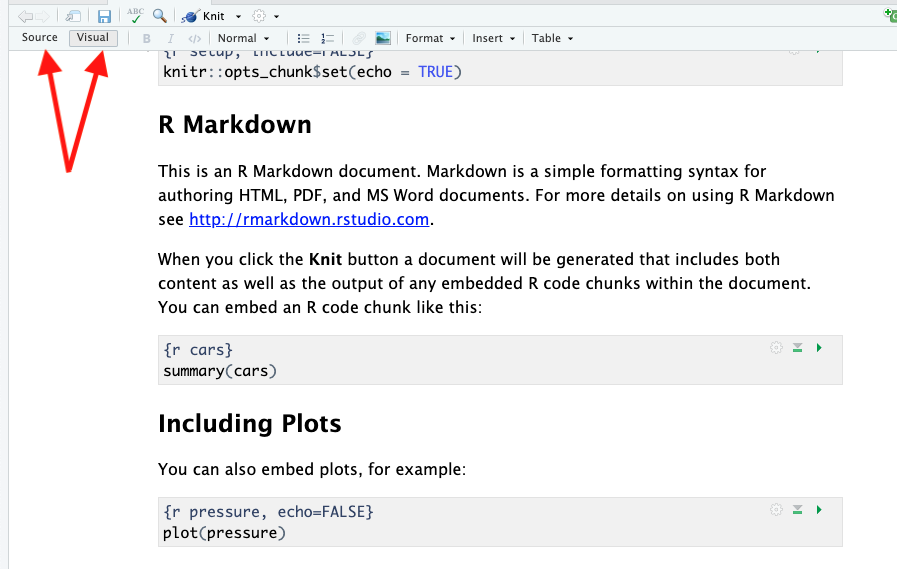
- In “Visual” mode, the document will look more like it’s final form. Headers will look like headers, bold will look bold, etc. I recommend that you use this mode most of the time.
- In “Source” mode, you will see all of the formatting codes and the document won’t be formatted. However, If you ever send me code, you must change it to “Source” mode before copying your code, because the formatting marks are important to include.
Before you move on, try toggling between the two modes to see how different the same document looks in each mode.
Knitting the documents
When you have finished a project, you would like to present a document that incorporates your code, output from your code (such as tables and figures) and text to explain what the analysis means. You need to “knit” the different parts of the file together.
- Click the little arrow next to the word ‘knit’.
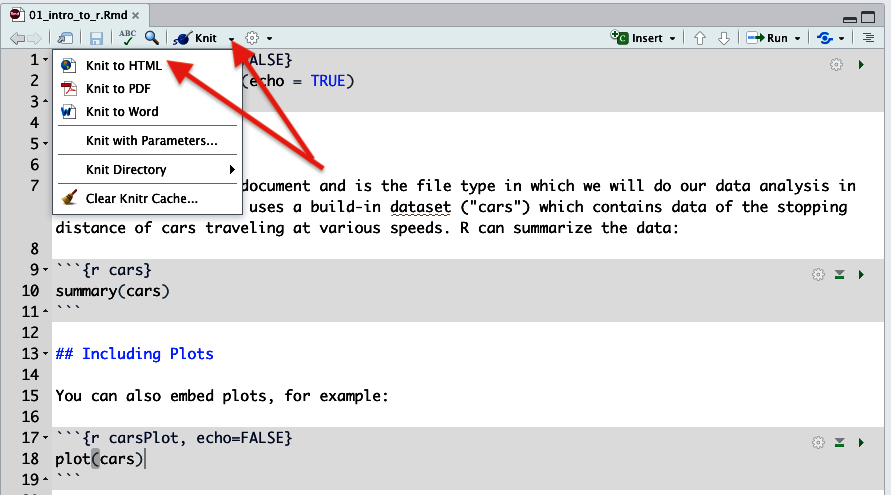
Figure 11. Knitting to produce an HTML document. Before you do this to turn in an HTML document, you must also select 'Clear Knitr Cache' from this same menu - Then click “Knit to HTML”. This will run all the code in the R Markdown file.
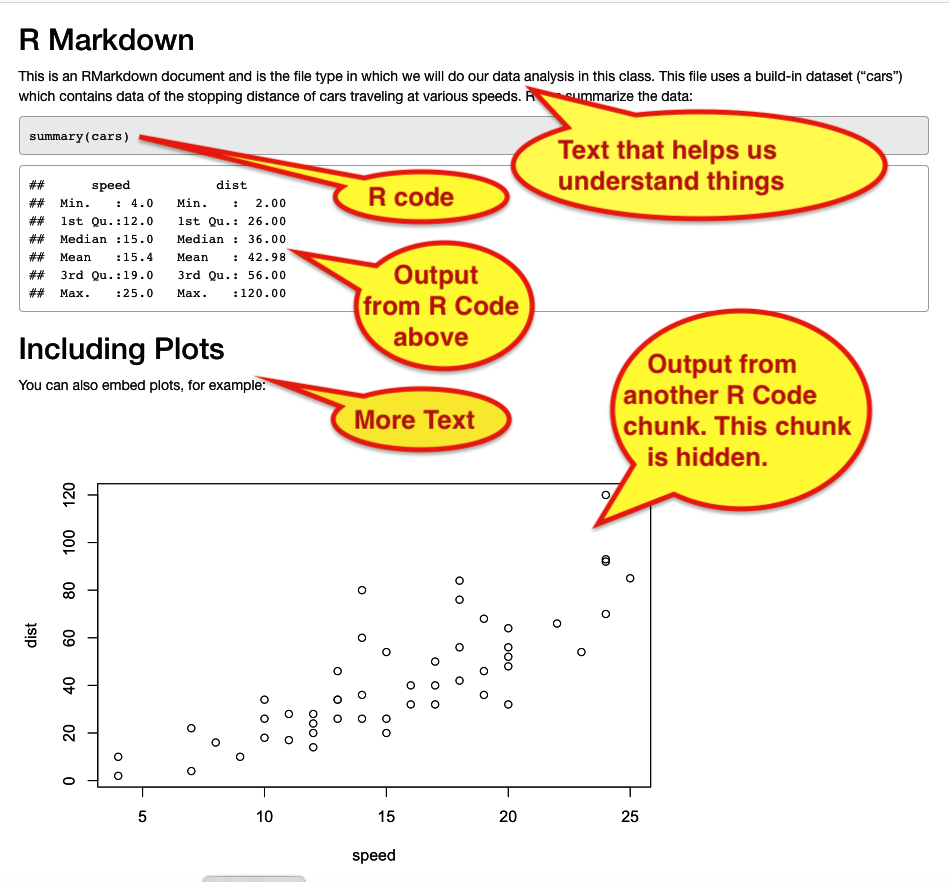
Figure 12. HTML output from knitting. - You will see an HTML file pop up. It has the text, code, and output (in this case, a graph), all ‘knitted’ together.
- in addition to the file that pops up, you will also see an .html file appear in the “Files” tab at the bottom-right.
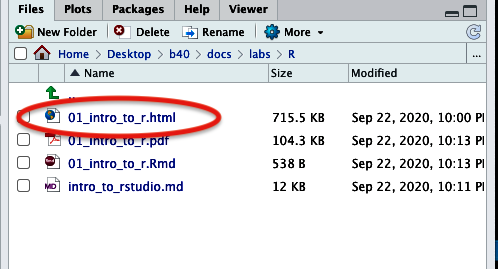
Figure 13. Knitted documents appear in lower right quadrant under 'Files' tab
If (when) you run into trouble
During the course of this semester, you will run into trouble and get stuck at some point. I can help!!! When this happens email me your entire .Rmd file either as an attachment or cut and paste it into your email. If you cut and paste, you must use “source” mode so that you also cut and paste all the formatting marks. If you don’t, you will simply be sending me a file that I cannot run, and I won’t be able to help you.
What to turn in
For this exercise, you only need to make a single change to the Rmd document. Usually, you will make changes in order to use your own data and analysis.
- If you haven’t done so already, download this R Markdown document and save it into a directory on your computer. I recommend that you set up a directory exclusively for Rmd documents for this class.
- Open the file and change the author name on the third line of the file to your own name.
- Save your file (you should do this periodically – it doesn’t save automatically like Google docs)
- Clear the knitr cache. To do so, click the triangle next to ‘Knit’ and select ‘Clear Knitr Cache…’.
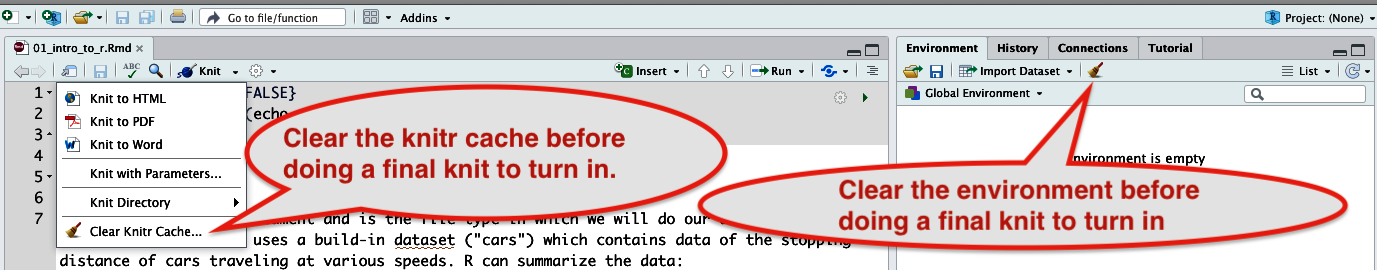
Figure 14. Clear the knitr cache and the environment as preparation before you do a final knit to generate the HTML file to turn in. - Clear your environment. Click the broom in the top-right quadrant. After, it should say “Your environment is empty”.
- Knit the document that you loaded (“intro_to_rstudio.Rmd”). This will create a .html file in the directory you are working, and it may also open a browser window.
- Save the resulting .html file to turn in. You can save it directly from your browser “file” menu if you have it open.
- Open the file in a browser (if it’s not already) to make sure it shows the results of your ‘data analysis’.
It is important that you clear your knitr cache and your environment before you prepare your document to turn in. If you don’t, you may get some experimental code or data that you played with but didn’t work.